March, 2013
Griffin has updated the 2011 direct2experts: http://direct2experts.org/IntegratedDemo
March, 2013
http://research.icts.uiowa.edu/polyglot/googleSearch.jsp
which interrogates one of three different configurations of Google CSEs. The first two interrogate the query interfaces of the 50-some direct2experts sites, either targeting people or just taking what comes back. The third covers the 61 CTSA sites linked off CTSACentral, plus CTSACentral itself.
Please also note that this is using the free interface, so we're capped a 100 queries a day, and only 10 results per query, so I'm not letting you past the first page of results…
April 10, 2013
The CTSAsearch prototype implementation of federated search is now available, using Linked Open Data published by members of the CTSA Consortium and other interested parties. To try it out, go to http://research.icts.uiowa.edu/polyglot
CTSAsearch currently indexes 41,367 persons and their 286,066 publications from Cornell, Florida, Iowa and Northwestern. Indexed content includes
- Persons and their associated properties (name, research statement, etc.)
- Academic Articles for each person with associated properties. Where DOIs or PMIDs exist additional information is added from MEDLINE, including
- Abstract
- Keywords
- MeSH terms
- Chemicals
- Genes
The CTSA federated search is a project of several servers being simultaneously searched and aggregated.
Information about the search from another site can be found at UIOWA.edu.
The page with a University of Florida theme. Installation ReadMeThe CTSAFederated search is a search ideally performed across several many servers simultaneously.
The "aggregatedsearch.php" pulls and displays the XML from the associated search sites. The "fs.php" code is designed to navigate the DOM of the VIVO search site and get the required data and provide it in the prescribed XML.
For this script to work all the parts are expected to be in the same directory.
the parts:
??_fs.php // ?? is the abbreviation of the instituion
at least one of (aggregatedsearch.php,aggregatedsearchVer2.php,aggsearch.php)
fsearchsites.xml //altered to point to each remote sites' ??_fs.php
simple_html_dom.php //library used to do the html DOM parsing.
The "??_fs.php" replaces the "fs.xml" with "??_fs.php?return=desc"
Variables that should be changed.
In ??_fs.php:
variables $school ,$name, and $icon are the school identifiers
variables $popdesc is the description of the population
In aggregatedsearch.php: (Ver2 is set to use the Icons in preference to the IFRAMES.)
variable $VivoHeader = Wrapper page HTML before results
variable $VivoFooter = Wrapper page HTML after results
variable $siteList = Location of fsearchsites.xml
aggsearch.php is geared towards being able to get its own wrapper.
Remember that any root reference (href="/blah.blah/stuff") will only work if they are on the same web server.
In aggsearch.php:
variable $wrapper = the HTML from the the page with the wrapper
variable $content = div to place the results into ('#contents' is id='contents' , '.contents' is class='contents')
variable $siteList = location of fsearchsites.xml
The aggregatedsearch.php uses fsearchsites.xml to get the location of the "??_fs.php" or the "fs.xml" file.
The folder "fs" has the current fs.xml files as seen from another box.
The versions of the fs.xml with the php files are set to work from "localhost".
the ??_fs.php file finds itself and returns the description.
fs.php is geared to reurn the vivo site results in the desired xml.
The ??_fs.xml sites point to the fs.php with the search data.
the flow of information can be shown as :
xml process:aggregatedsearch.php --calls-->fsearchsites.xml --to find--> ??_fs.xml --to call--> fs.php?(proper values) --to get--> search data --displayed--> aggregated search page.
php process :aggregatedsearch.php --calls-->fsearchsites.xml --to find--> ??_fs.php?return=desc --to call-->??_fs.php?return=xml&querytext=TERM --to get-->search data --displayed--> aggregated search page.
The other network sites will bet told where to find the ??_fs.php which gives the description to call ??_fs.php to get the xml of the search results.
update 2-25-2011:
The aggregatedsearch-UFsegmented.php is geared to use AJAX asynchronous loading to make the page faster.
To facilitate the calls a new php file resultsearch.php which takes the description location and the term to make the row of the table.
resultsearch.php is called on for each site in question, each instance does the search as another page request within the browser.
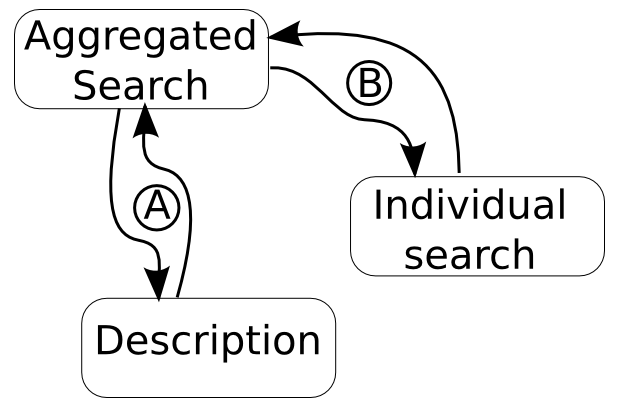
Data flowThe following is a brief summary of the flow of data. |
- The Aggregated search receives the query term
- Stage A:
- The Aggregated search sends requests for the descriptions
- The search then retrieves the descriptions of the sites.
- Stage B:
- The query term is sent to the site from the description
- The count, site, and preview site information is sent back. |
|
Examples
An example "fs.xml" or the return from "fs.php?return=desc"
<?xml version='1.0'?> <site-description> <name>University of Florida</name> <icon>http://www.vivoweb.org/files/logos/partner_institutions/ufl_logo.gif</icon> <aggregate-query>http://vivotest.ctrip.ufl.edu/ctsa/test/uf_fs.php?return=xml&querytext=</aggregate-query> </site-description>
The icon field is optional but was found useful for formats where an icon for the site would be more acceptable.
An example of the "fsearchsites.xml" which details the location of the xml site descriptions. This file's contents are the sites visited and the order to display them in.
<partnersites> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/uf_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/cu_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/iu_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/wustl_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/psm_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/weill_fs.xml</description-site-URL> <description-site-URL>http://vivotest.ctrip.ufl.edu/ctsa/fs/scripps_fs.xml</description-site-URL> <description-site-URL>http://staging.connects.catalyst.harvard.edu/profileslabs/fs.xml</description-site-URL> <description-site-URL>http://research.icts.uiowa.edu/polyglot/FS.xml</description-site-URL> <description-site-URL>http://irt-dev.stanford.edu/profiles-federated-pilot/FS.xml</description-site-URL> <description-site-URL>http://profiles.ahc.umn.edu/direct.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/einstein/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/jhu/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/maryland/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/mcg/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/miami/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/mmc/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/mskcc/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/ohsuV3/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/uab/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/ucdavis/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/uic/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/umichigan/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/uthscsa/FS.xml </description-site-URL> <description-site-URL>http://www.experts.scival.com/wayne/FS.xml </description-site-URL> </partnersites>
AJAX implementation
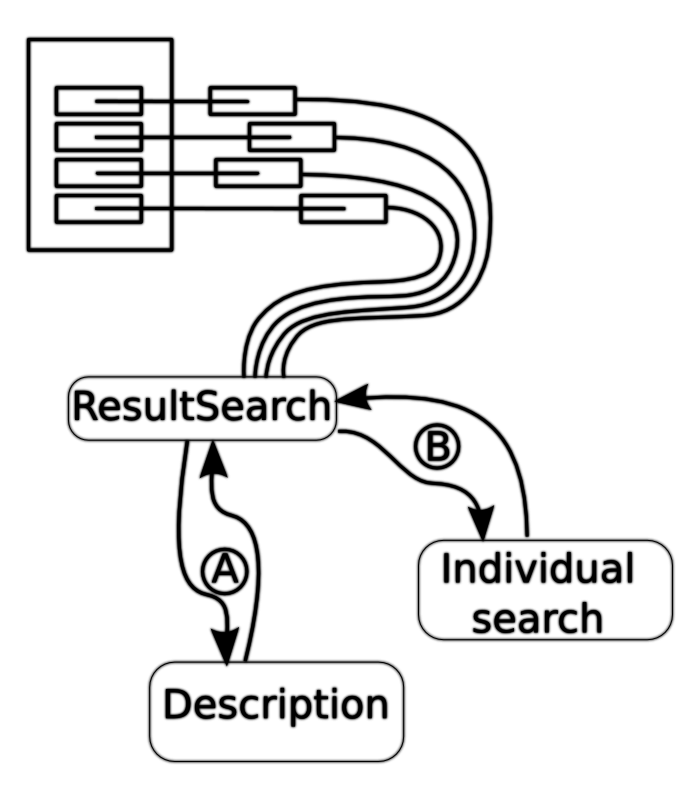
In the most recent version (aggregatedsearch-UFsegemented.php) of the search each of the page results are loaded asynchronously.
The aggregated search does a series of asynchronous calls to resultsearch.php. This resultsearch.php returns the significant information once it is available.
Data flow
The following is a brief summary of the flow of data.
- The Aggregated search receives the query term
- The Aggregated search calls resultsearch with the description URL and term
- Stage A:
- The resultsearch sends requests for the descriptions
- The search then retrieves the descriptions of the sites.
- Stage B:
- The query term is sent to the site from the description
- The count, site, and preview site information is sent back. |
|