Deprecated. This material represents early efforts and may be of interest to historians. It doe not describe current VIVO efforts.
Deprecated. This material represents early efforts and may be of interest to historians. It doe not describe current VIVO efforts.
Exercise: Finding VIVO Data with the University of Florida’s Public SPARQL Endpoint
This exercise was developed by Brian Lowe for the 2013 Introduction to VIVO Workshop
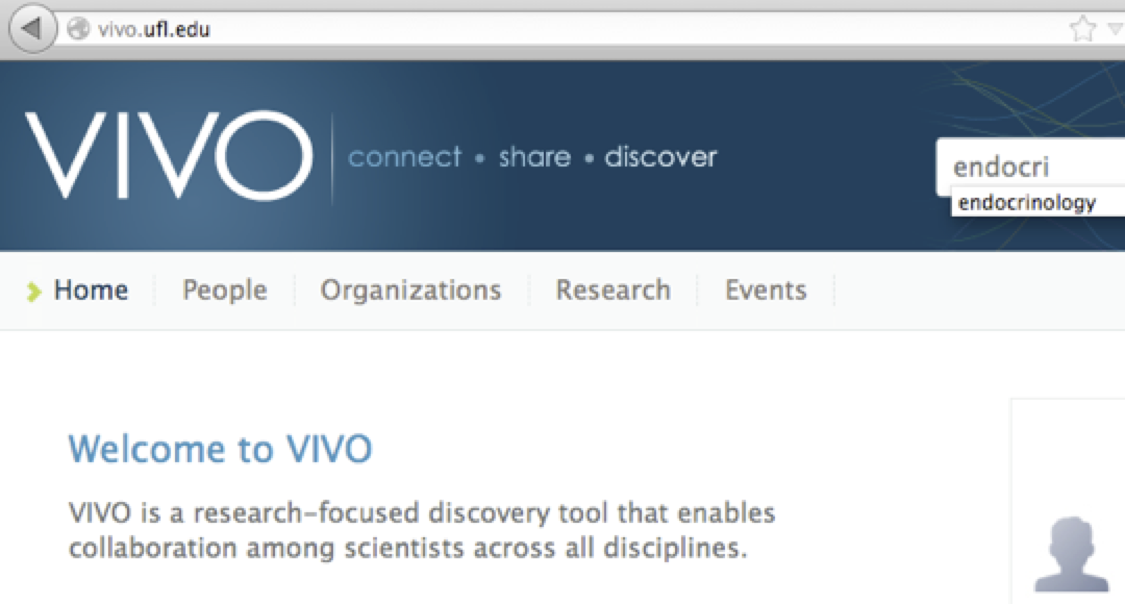
1. Visit http://vivo.ufl.edu/
2. Search for ‘endocrinology.’
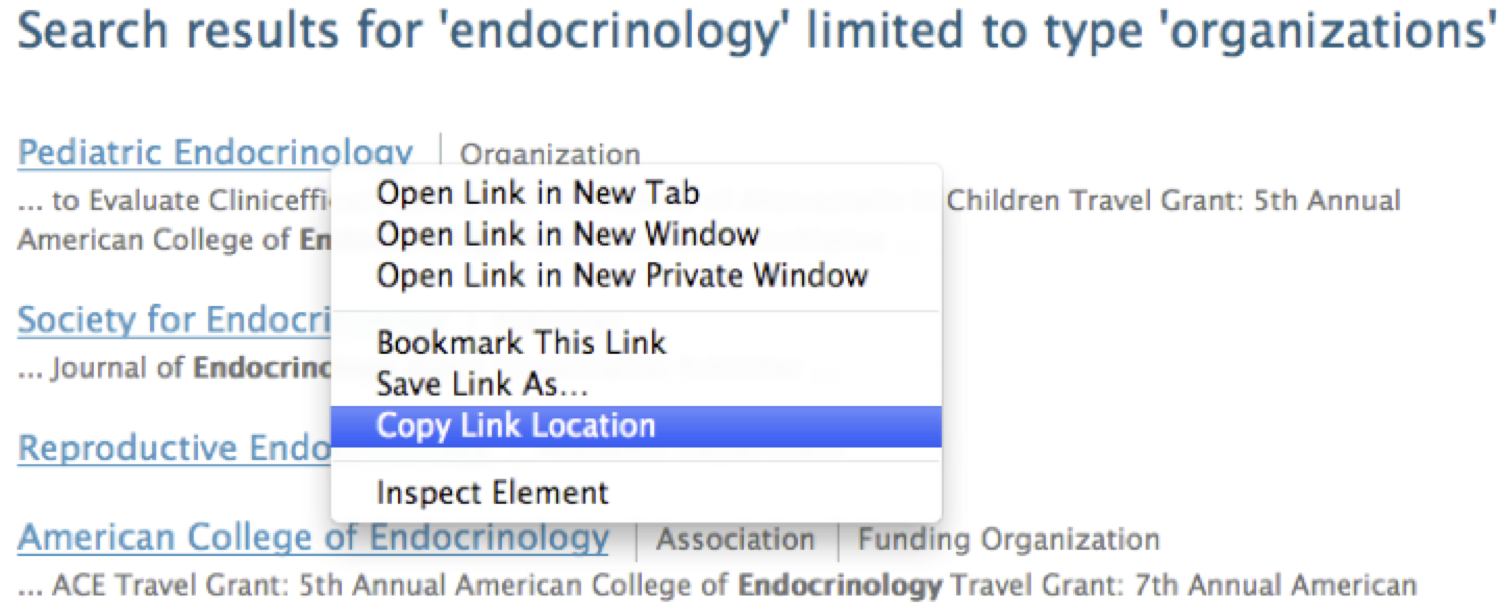
3. At the right side of the page, under “Display only,” click “organizations” to show only search results that are organizations.
4. Let’s inspect the linked data available for the first search result. Right-click and copy its Web address.
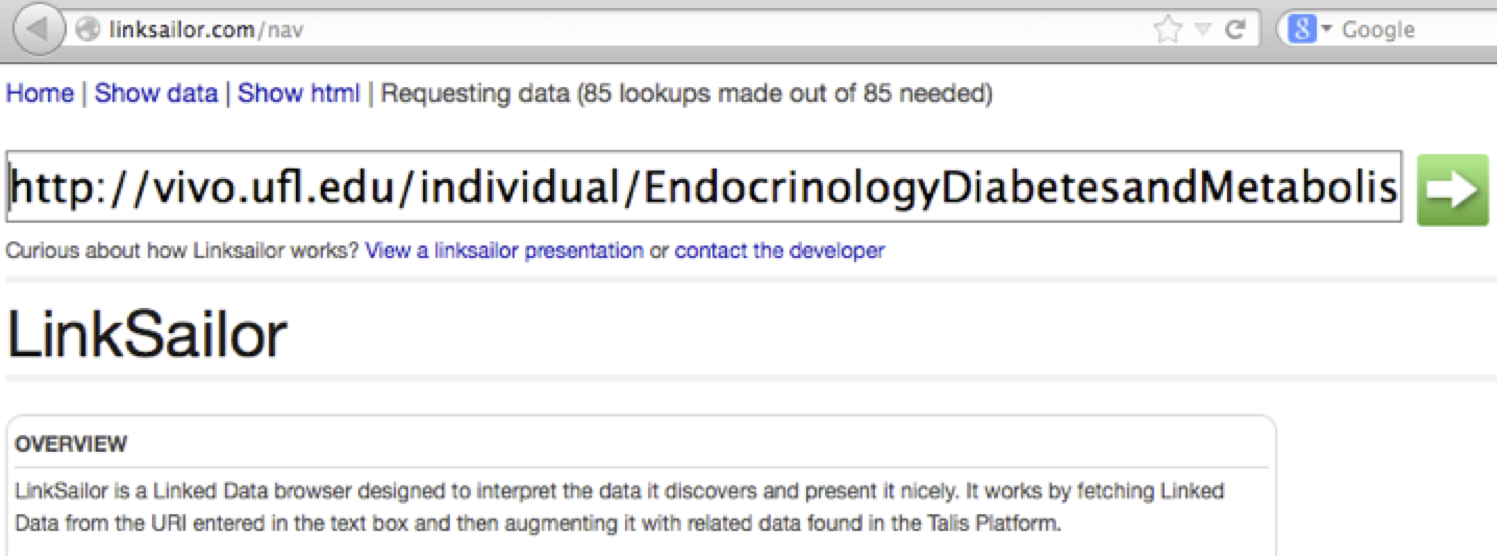
5. Visit http://linksailor.com/ and past the address into its search box.
6. Click the green arrow. When the next page appears, click “Show data” in the upper left-hand corner. You should see Linksailor’s rendering of the raw data:
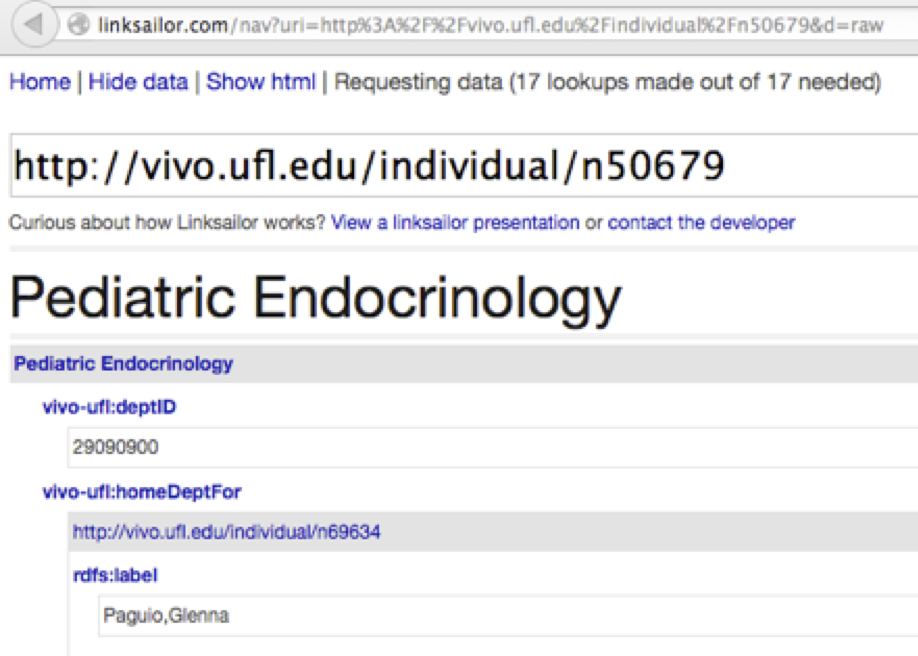
7. Scroll all the way to the bottom, where you should see some basic triples directly related to this department:
The facet control on the search results page was able to identify this search result as an organization because the linked data specifies that it has an rdf:type value of http://xmlns.com/foaf/0.1/Organization. The search was able to return this resource in a search for ‘endocrinology’ because the linked data specifies an rdfs:label value containing that string.
8. The SPARQL query language lets us query the structured semantic relationships directly, instead of relying on text matches. To begin, visit http://sparql.vivo.ufl.edu/.
9. Click the first link, “Control Panel.”
10. Click the “Select” button.
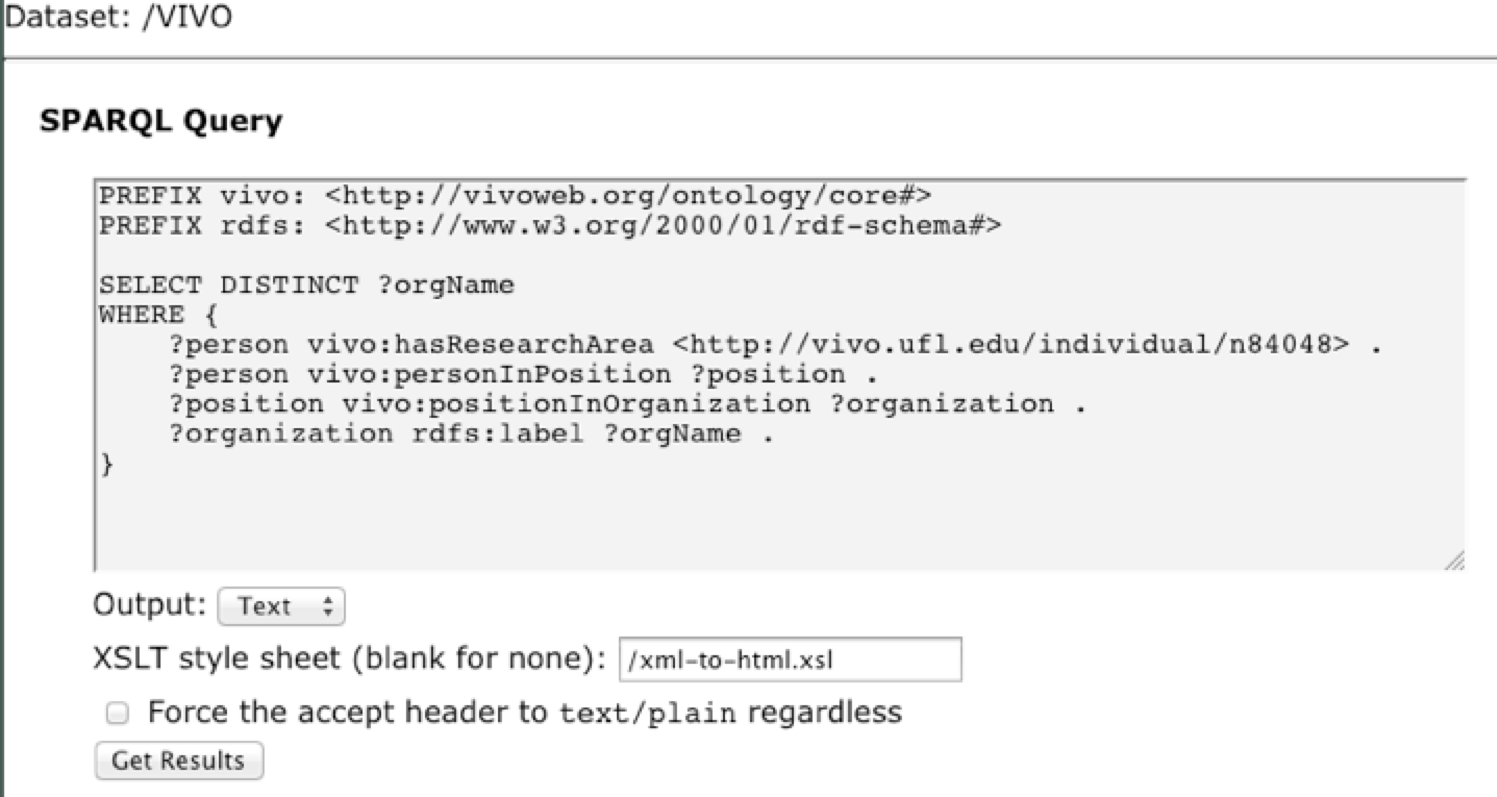
11. Let’s write a query that finds organizations whose members do research on endocrinology. Instead of searching on a text string, we’ll use the URI of a SKOS Concept for endocrinology, http://vivo.ufl.edu/individual/n84048. Our query will find people who are specified as having this concept as a research area, and additionally ask for the organizations in which they hold positions.
Enter the following query into the text box:
PREFIX vivo: <http://vivoweb.org/ontology/core#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT DISTINCT ?orgName
WHERE {
?person vivo:hasResearchArea <http://vivo.ufl.edu/individual/n84048> .
?person vivo:personInPosition ?position .
?position vivo:positionInOrganization ?organization .
?organization rdfs:label ?orgName .
}
This is what it should look like in Fuseki:
12. Select “Text” in the Output dropdown list and then press the “Get Results” button. After a few seconds you should see some results.
13. Now let’s augment the query with additional relationships. We’ll follow a longer chain of relationships from people who have endocrinology as a research area in order to discover the research areas of their coauthors.
Press the back arrow and paste this query into the text box:
PREFIX vivo: <http://vivoweb.org/ontology/core#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT DISTINCT ?researchAreaName
WHERE {
?person vivo:hasResearchArea <http://vivo.ufl.edu/individual/n84048> .
?person vivo:authorInAuthorship ?authorship .
?authorship vivo:linkedInformationResource ?doc .
?doc vivo:informationResourceInAuthorship ?authorship2 .
?authorship2 vivo:linkedAuthor ?coauthor .
?coauthor vivo:hasResearchArea ?researchArea .
?researchArea rdfs:label ?researchAreaName .
FILTER(?authorship != ?authorship2)
}
This query will return some different concept names:
14. By replacing the concept URI http://vivo.ufl.edu/individual/n84048 with different concept URIs, we can find different sets of organizations and coauthor research areas. How can we find different URIs to try?
Try this query. It may take a few seconds to return the list.
PREFIX vivo: <http://vivoweb.org/ontology/core#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
SELECT DISTINCT ?concept ?conceptName
WHERE {
?concept a skos:Concept .
?concept rdfs:label ?conceptName
}
<end of exercise>